Biological neurons
A biological neuron (nerve cell) is an electrically excitable cell that processes and transmits information through electrical and chemical signals. A chemical signal occurs via a synapse, a specialized connection with other cells. Neurons connect to each other to form neural networks. Neurons are the core components of the nervous system, which includes the brain, spinal cord, and peripheral ganglia. There are different types of neurons: sensory neurons, motor neurons and interneurons.
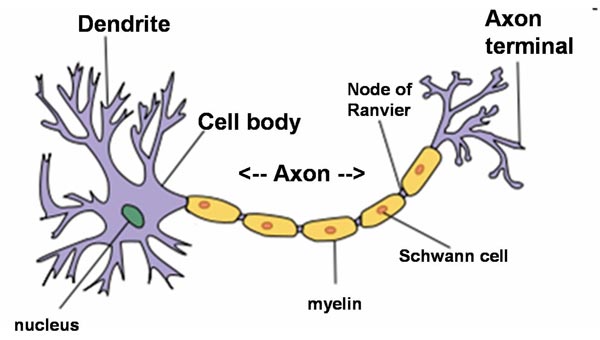
A typical neuron possesses a soma (perkaryon or cyton = cell body with nucleus), dendrites and an axon. Neurons do not undergo cell division.
Neuron (Wikipedia)
Dendrites are thin structures that arise from the cell body, branching multiple times and giving rise to a complex dendritic tree. An axon is a special cellular extension that arises from the cell body and travels for long distances (as far as 1 meter in humans). The cell body of a neuron gives rise to multiple dendrites, but never to more than one axon, although the axon may branch hundreds of times before it terminates. The axon terminal contains synapses, specialized structures where neurotransmitter chemicals are released to communicate with target neurons. At the majority of synapses, signals are sent from the axon of one neuron to a dendrite of another, however there are a lot of exceptions.
All neurons are electrically excitable, maintaining voltage gradients across their membranes by means of metabolically driven ion (sodium, potassium, chloride, calcium) pumps. Changes in the cross-membrane voltage can alter the function of voltage-dependent ion channels. Each time the electrical potential inside the soma reaches a certain threshold, an all-or-none electrochemical pulse called an action potential is fired, which travels rapidly along the cell’s axon, and activates synaptic connections with other cells when it arrives.
Artificial neurons
An artificial neuron is a mathematical function conceived as an abstraction of biological neurons. The artificial neuron receives one or more inputs (representing the dendrites) and sums them to produce an output (representing the axon). Usually the sums of each node are weighted, and the sum is passed through a non-linear function known as an activation function or transfer function.
The first artificial neuron was the Threshold Logic Unit (TLU) first proposed by Warren McCulloch and Walter Pitts in 1943. This model is still the standard of reference in the field of neural networks and called a McCulloch–Pitts neuron. However, artificial neurons of simple types, such as the McCulloch–Pitts model, are sometimes characterized as caricature models, in that they are intended to reflect one or more neurophysiological observations, but without regard to realism.
In the 1980s computer scientist Carver Mead, who is widely regarded as the father of neuromorphic computing, demonstrated that sub-threshold CMOS circuits behave in a similar way to the ion-channel proteins in cell membranes. Ion channels, which shuttle electrically charged sodium and potassium atoms into and out of cells, are responsible for creating action potentials. Using sub-threshold domains mimicks action potentials with little power consumption.
At the Neuromorphic Cognitive Systems Institute of Neuroinformatics of the University of Zurich and ETH Zurich, a research group leaded by Giacomo Indiveri is currently developing, using the sub-threshold-domain principle, neuromorphic chips that have hundreds of artificial neurons and thousands of synapses between those neurons.