Last update : October 20, 2016
Note : the new versions released since spring 2016 have not yet been tested. It will be done in the coming days.
Introduction
This contribution refers to my recent updated post about DICOM image viewers. Mobile DICOM viewers are becoming more and more popular. They are very useful in the context of medical applications in the cloud.
Most web viewers can be used on mobile equipments (tablets, smart phones, …), but the focus of this post are native applications designed for Apple iOS or Android devices. DICOM Web viewers are handled in a separate contribution. In the present list I considered only those applications that have been updated after January 2014, with some exceptions.
Test Scenario
To test the selected apps, I used Orthanc (Windows versions 0.9.5 and 0.9.6) as PACS server. To have realistic DICOM samples, I uploaded some DICOM studies from me and from family members relative to exams done in the past in hospitals in Luxembourg and France.
To set up a communication between a DICOM viewer and the Orthanc server, the viewer should allow to define the following parameters in the settings :
- Remote AET
- Remote Hostname/IP
- Remote port number
- Local AET
- Local port number
- Protocol : C-MOVE
The local IP is the address of the used mobile device.
If no settings are available for the port numbers, the port number of the server can be added to the remote IP address. The standard port number 104 can be tried as a first guess for the settings of the viewer. If no local AET settings are provided, the iPad name can be tried as a first guess. Another possibility is to use a DICOM network analyser like DVTk to detect unknown parameters. If the C-MOVE protocol is not supported by the viewer, the WADO or C-GET protocols can be an alternative.
In the presentation of the selected mobile viewers, I generally used the terminology published by the developers, even if I disagree with their claims. In the case of apps that worked as expected with the Orthanc server, I added some screenshots to show the look-and-feeling of the viewer.
Purpose of the evaluation
You may ask what is the purpose to evaluate proprietary closed source applications in an open source environment. I started the evaluation in mid-2015 in the context of the development of an educational tool for radiologists, called RadioLogic. The reasons were various :
- to familiarize with the DICOM technologies (learning by doing)
- to compare the performance of native apps with HTML5 web apps on tablets and phones
- to explore the different types of graphical user interfaces to create a best-of-breed GUI for the RadioLogic project
- to examine the assets of viewers certified / cleared by FDA or CE
- to test features that are not (yet) available on HTML5 web apps
iOS Applications

Mobile DICOM viewers (icons) arranged in a folder on the iPad Desktop
The following DICOM viewer apps, which are currently available at the AppStore, have been tested. They are arranged and listed in alphabetical order :
1. aycan mobile
aycan mobile is an iPad app which allows to receive and display DICOM images in an easy, secure and fast way. Developed by aycan Digitalsysteme GmbH, the current version 1.08.000 was updated on July 22, 2015. The viewer works only with the aycon DICOM workstation, which is based on Osirix. A demo server with ID and password “aycan” allows to explore some demo studies. A subscription is required to use all features of the the ayMobile communication service. A free 30-days-trial subscription can be ordered on the aycan website.
2. DICOM-AVC
DICOM-AVC is available for iPad and iPhone. The current free version is 2.2 has not been updated since July 12, 2012. It has been developed by the The New Medical Product Server LLC who claims that DICOM-AVC is a complete real-time collaborative imaging and videoconferencing tool which was once the number one downloaded app on the AppStore. To use the app, registration of an account is required, which can be upgraded to a premium account for 19.99$ per year. Today it’s displayed as black screen on my iPad.
3. DICOM Mobile
Dicom Mobile is a simple but powerful DICOM viewer for iPhone and iPad, developed by Luigi Orso. The current version is 1.4.0, updated on June 3, 2015. The price is 2.99$, with additional in-app purchases for extensions. The developer provides support on his website. DICOM files can be loaded from a DropBox account or from a Web server, but not from a PACS server.
DICOM Mobile Main Menu
DICOM Mobile Dropbox Panel
4. DICOM Viewer HD
The free version 1.0 of DICOM Viewer HD was released on July 4, 2014, by Wingspan Xi’an Electronic Technology Co., Ltd from China. I was not able to evaluate the app, the login or registration in the “User Manageer” always fails.
5. DICOM IR
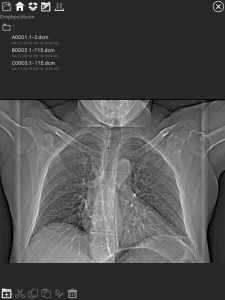
DICOM IR is one of the easiest and most intuitive 2D DICOM Image viewers for iOS. DICOM IR can browse any standard PACS server. DICOM IR is developed by Pietro Zuco, a freelance mobile developer based in London/Tokyo/Barcelona. Pietro Zuco has a solid experience developing 2D games, utilities, educational and entertainment applications on iOS and OSX systems. Version 1.0 of DICOM IR was released on July 25, 2015 and is free for a limited time. The app supports C-ECHO, C-FIND and C-MOVE queries.
DICOM IR has a very clean GUI, an excellent user guide, a perfect configuration panel, a performant viewer and it works as expected with the Orthanc server. My compliments to this author.
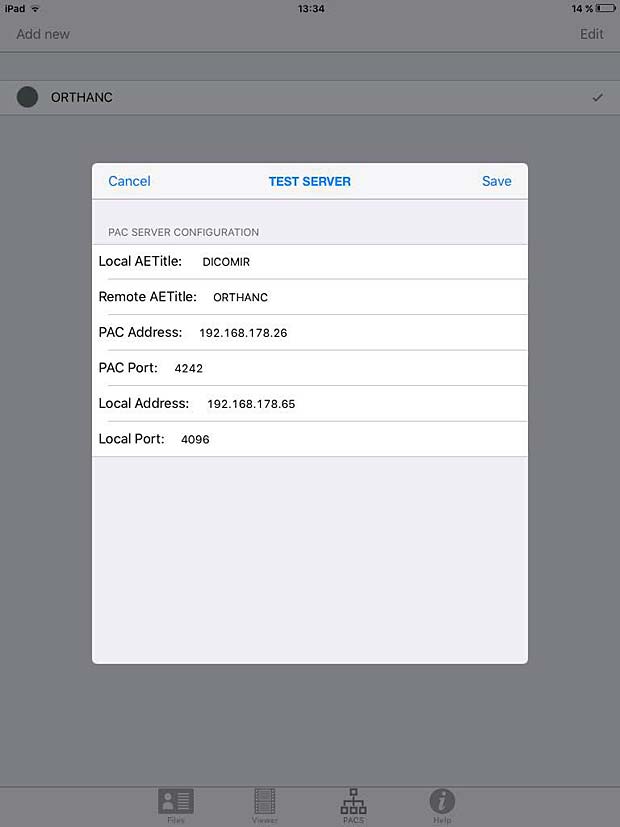
DICOM IR PACS Server Configuration Panel
DICOM IR User Guide
DICOM IR search panel

DICOM IR Metadata Info panel
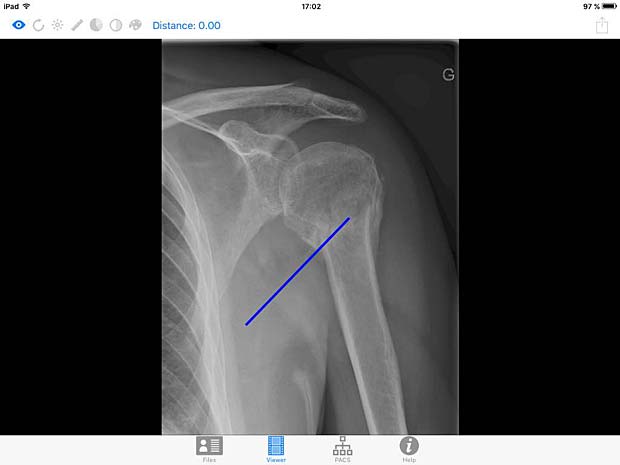
DICOM IR image viewer panel with line tool
6. iDO Viewer
The iDO medical image viewer is quickly becoming the next generation in mobile medical image viewing by combining multi-modality imaging with individualized physician workflow. Developed by EBM Technologies, Incorporated, the current version 1.1.4 was updated on March 30, 2016. This application requires the EBM iDO server, demo servers to view sample data and basic features of the app are available and can be selected in the settings panel.
7. imageFlow.mobile
imageFLOW.mobile is the best-in-class mobile application for medical imaging PACS users on iOS devices. imageFLOW is developed by imagitive GmbH. The current free version 2.0, based on Imagitive’s DaVinci framework, was updated on November 25, 2015. A DemoServer mode to explore some DICOM studies is enabled by default. A PACS server can be configured in the general iOS settings, nevertheless access trials failed with a “Server Authentication” error message.
8. iMango
iMango is the iOS adaptation of the famous Mango DICOM viewer developed at the Research Imaging Institute (RII), an Organized Research Unit (ORU) of the University of Texas Health Science Center at San Antonio. It works on iPad’s. The current version 1.3 was updated on June 10, 2015. The price is 14.99$. A PACS server can be configured in the general settings and the app works as expected with the Orthanc server. iMango also supports the WADO protocol.
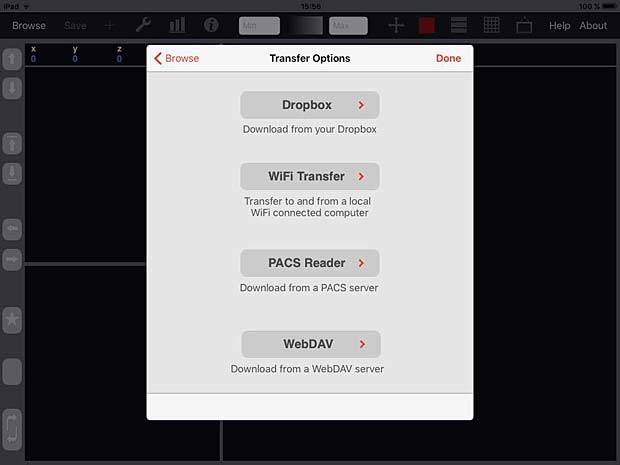
iMango Transfer Options Panel
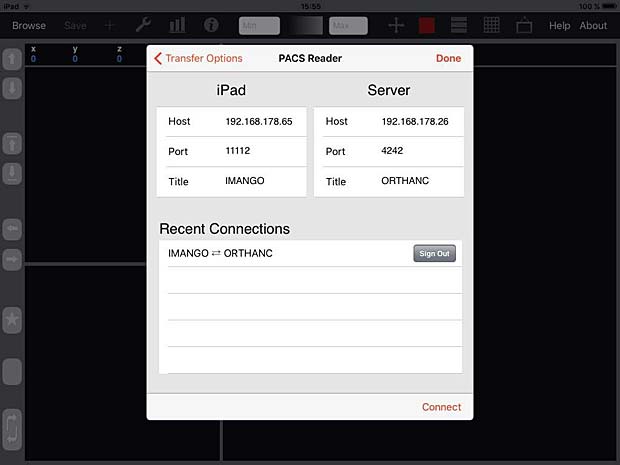
iMango Server Configuration Panel
iMango User Guide
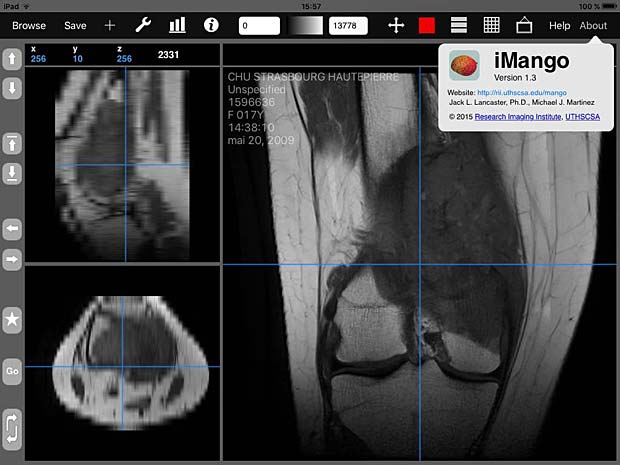
iMango image viewer with about panel
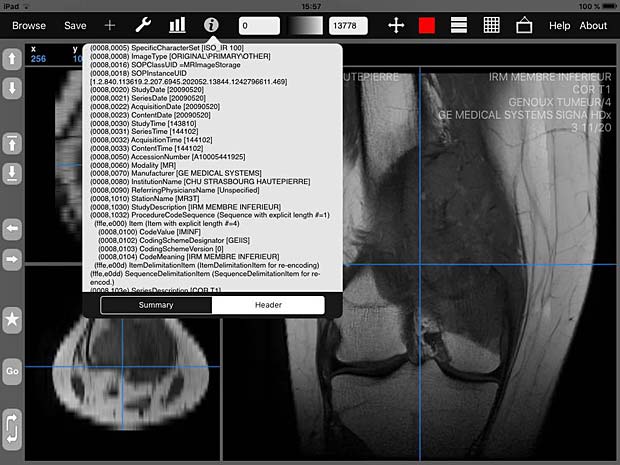
iMango image viewer with metadata info panel
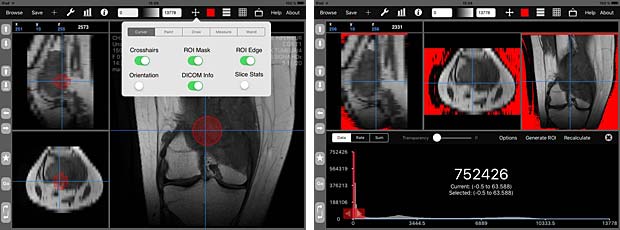
iMango image tools
9. iPaxera
iPaxera is one of the fastest and easy to use PACS viewing applications specifically designed for viewing and navigating studies with your iPad, iPhone, and iPod. iPaxera is developed by Paxeramed Corp, a world leading medical imaging solution developer based in Boston, that develops a full range of medical imaging solutions. The current free version 3.10 was updated on May 4, 2016. A PACS server can be configured in the “Add Node” panel. Ipaxera works as expected with the Orthanc server. The app also supports the WADO protocol.
Onn Akbar Ali reported that Ipaxera can display multiframe Jpeg images ( such as angiography ) quite well.

iPaxera login panel
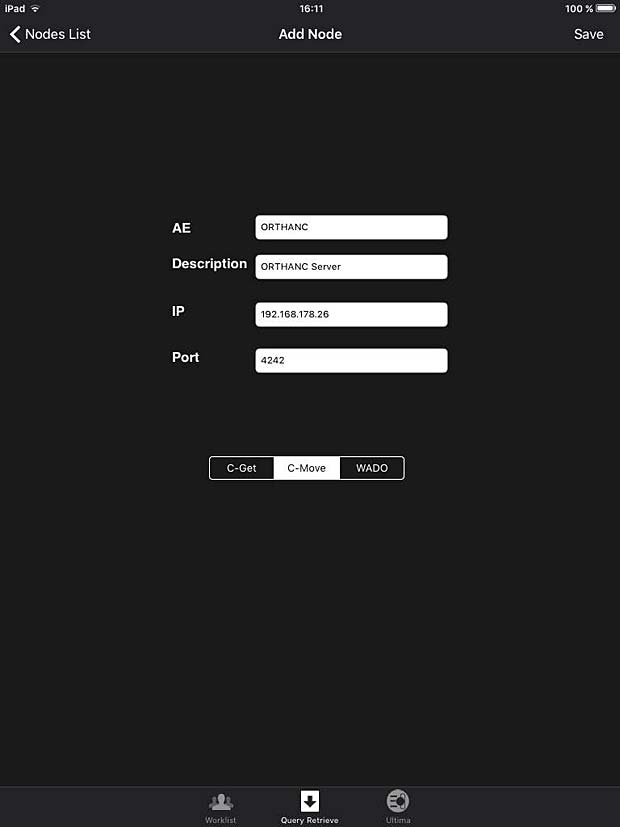
iPaxera PACS server configuration panel
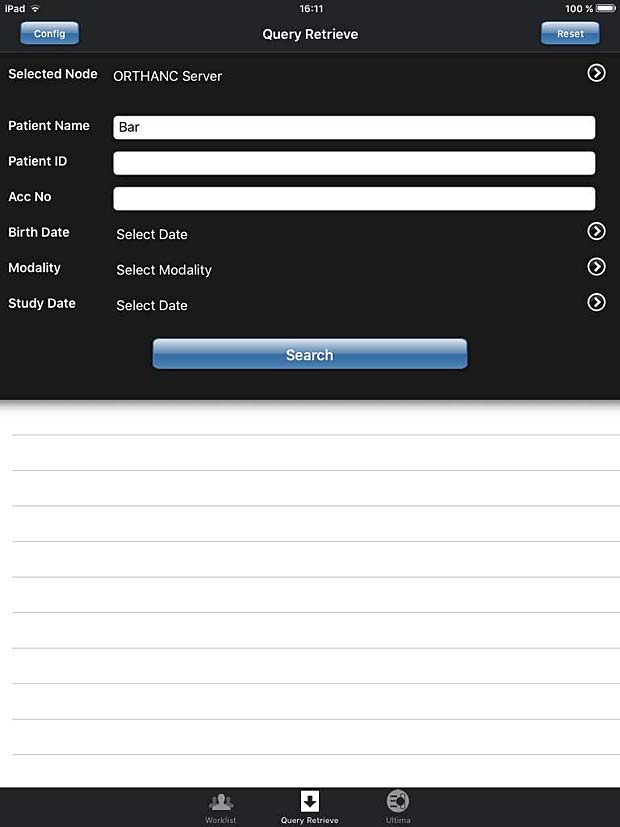
iPaxera search panel

iPaxera worklist panel

iPaxera image viewer with measure tool and series list panel
10. iTA View
iTA VIEW is a viewer for DICOM and DICOM RT objects developed for iPad and dedicated to radiotherapy. The app is developed by Tecnologie Avanzate TA srl in Italy. The current version 1.2.0 was updated on March 8, 2016. The app works with the Orthanc server. To transfer studies without limitations, an upgrade to the full version is required. The price for the upgrade is 29.99 EUR.

ITA View PACS server configuration panel
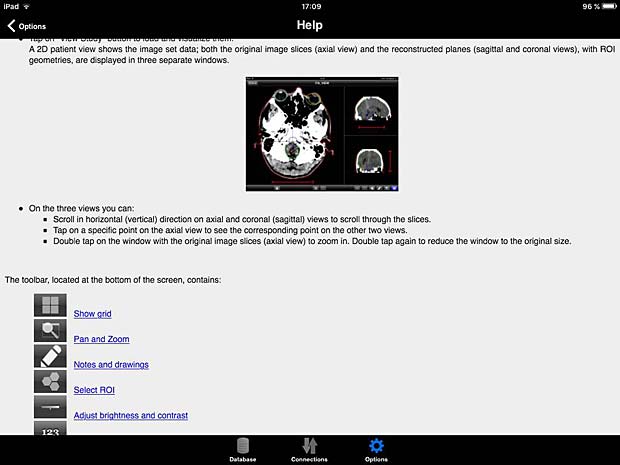
ITA View User Guide
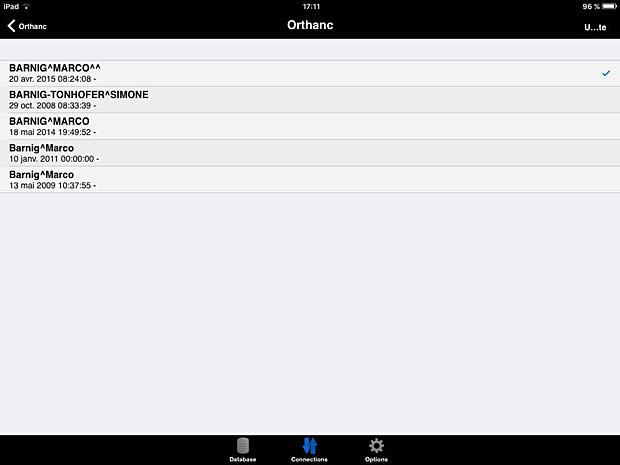
ITA View worklist panel
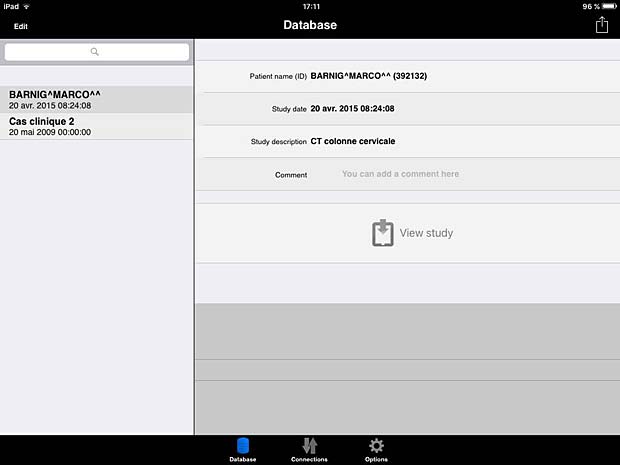
ITA View database panel
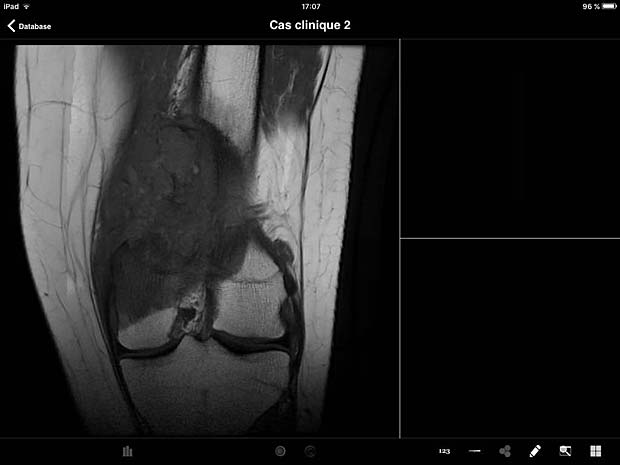
ITA View image viewer
11. Jack Imaging Medical Image Viewer
Jack Imaging Medical Image Viewer is a medical image viewer for the iPad that makes use of nephosity’s patent pending technology, the latest in medical imaging innovation. It supports reading the DICOM format and the most common imaging modalities. The viewer is developed by Nephosity Inc. The current version 3.0.5 was updated on January 25, 2016. The prize is 0.99$. The viewer works only with the Jack Imaging platform which stores the DICOM data in the cloud. Currently Amazon’s cloud infrastructure is used to provide the best performance and highest security levels. During a beta phase the Jack Imaging platform accounts are offered for free. On 4th generation iPads the Jack Imaging viewer offers FDA cleared capabilities.
11. Keiron
Keiron is an innovative App that allows you to view, annotate and analyse radiographic images stored in the DICOM standard. The current free version 1.0 was released on May 4, 2015. Keiron is developed by Digital Video S.p.A., the creator of Toonz. A PACS server can be configured in the settings, but only the ECHO and C-FIND queries are working with the Orthanc server. The “download” button to load DICOM instances is disabled.
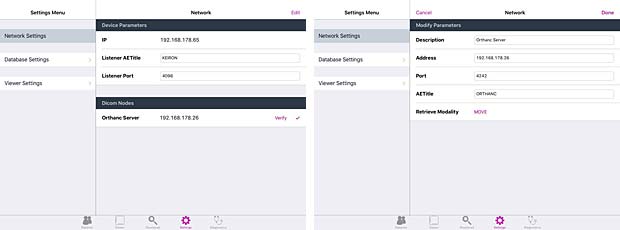
Keiron server & client configuration panels
Keiron viewer settings panel
Keiron worklist panel
12. LEADTOOLS MedicalWebViewer
The LEADTOOLS Medical Web Viewer is a zero footprint solution available for iPad and iPhone, developed by Leadtools Technologies Inc. The current free version 3.0 was updated on May 27, 2016. The viewer works only with LEAD’s web servers so you can quickly access the features of the LEADTOOLS Medical Web Viewer Framework SDK with the default settings.
13. loDicom Viewer
loDicom Viewer allows you to view, archive and transfer 2D medical images in DICOM format. Developed by Luigi Orso, the current version 4.1.1 was last updated on July 30, 2014. The price is 12.99$. The developer is also the author of the DICOM Mobile viewer. He provides support on his website, which is not useless, because the app crashes systematically on my iPad. ECHO tests with the Orthanc server are succesful, but a C-FIND query leads to a crash. I was only able lo load DICOM files via a DropBox. The GUI is not very intuitive, the user guide is not very helpful.
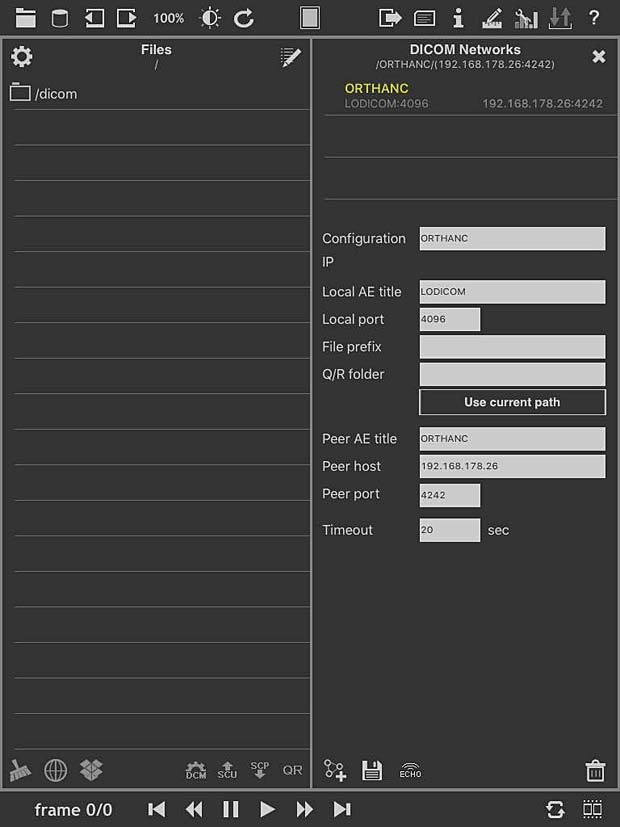
loDICOM Viewer PACS server configuration panel
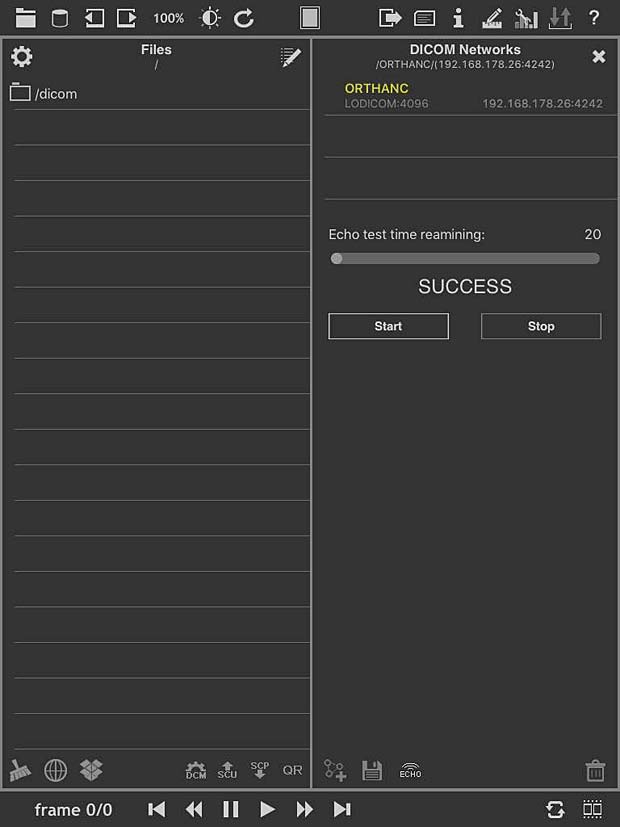
loDICOM Viewer ECHO Test panel
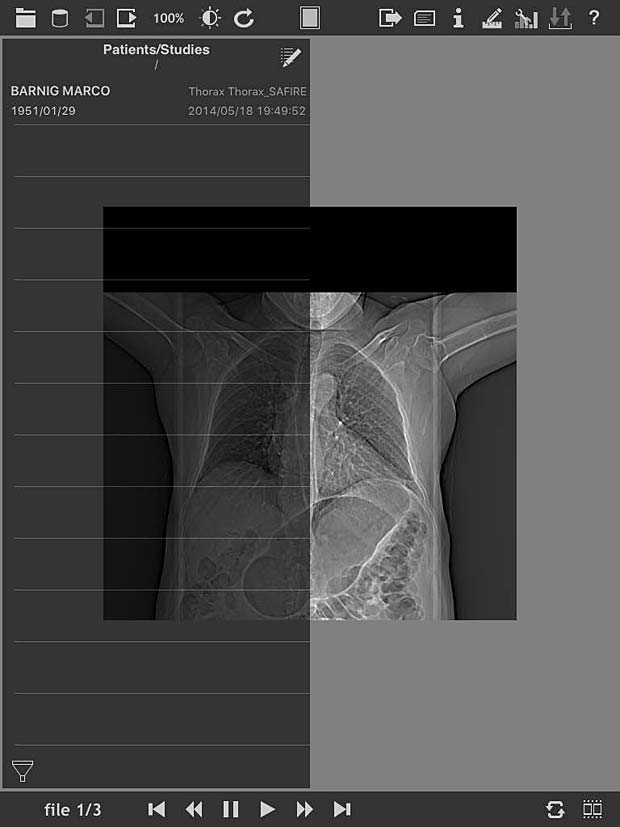
loDICOM Viewer image panel

loDICOM Viewer user guide
14. MedFilm
MedFilm is a fast dicom viewer developed by Fuli Wu, a researcher at the Zhejiang University of Technology in Hangzhou, China. The app is available for iPad and iPhone. The current free version 4.25 was updated on June 1, 2016. A PACS server can be configured with support of the WADO protocol. The C-MOVE protocol is not supported.
MedFilm DICOM file and image panels
15. Med View
Med View is developed by Matthias Rudolph and available for iPad and iPhone. The current free version 1.1 was updated on October 23, 2015. DICOM files can be loaded from the iTunes file sharing service or from FTP servers.
16. Mobile MIM
The Mobile MIM software program is used for the viewing, registration, fusion, and/or display for diagnosis of medical images from various modalities. Developed by MIM Software Inc., the current version 4.0 was updated on July 26, 2016. Image data can only be transferred to the device by using MIMcloud or MIM 5.1+ workstation software.
17. mRay
mRay, developped by mbits, is available for iPad and iPhone. The current free version 4.1.6 was updated on October 4, 2016. mRay is a CE certified and approved medical product. To be able to receive images in mRay from your PACS, the mRay Server Software must be installed, which can be used free of charge for an evaluation period.
18. MyXray
MyXray is an easy and intuitive app that will let you see your x-ray files as a real doctor. The free version 1.0 was released on September 8, 2015. The app is developed by Pietro Zuco, the author of the outstanding DICOM IR app. MyXray is a lite version of DICOM IR without support of DICOM communications.
19. Nuance PowerShare
Nuance PowerShare Mobile gives you instant access to your medical images and reports stored on the Nuance PowerShare Network using your iOS device. The free version 5.5.10 was updated on August 16, 2016. I was not able to load this app from the AppStore.
20. OsiriX HD
OsiriX HD is the iOS adaptation of the famous OsiriX DICOM viewer, running on Mac OSX, which is considered as a reference for DICOM image viewers. It works on iPhone’s and iPad’s. The current version 4.2 was updated on October 14, 2016. The price is 49,99$, with additional inline sales. Osirix HD can be configured in the general settings. The app works as expected with the Orthanc server and supports also the WADO protocol.
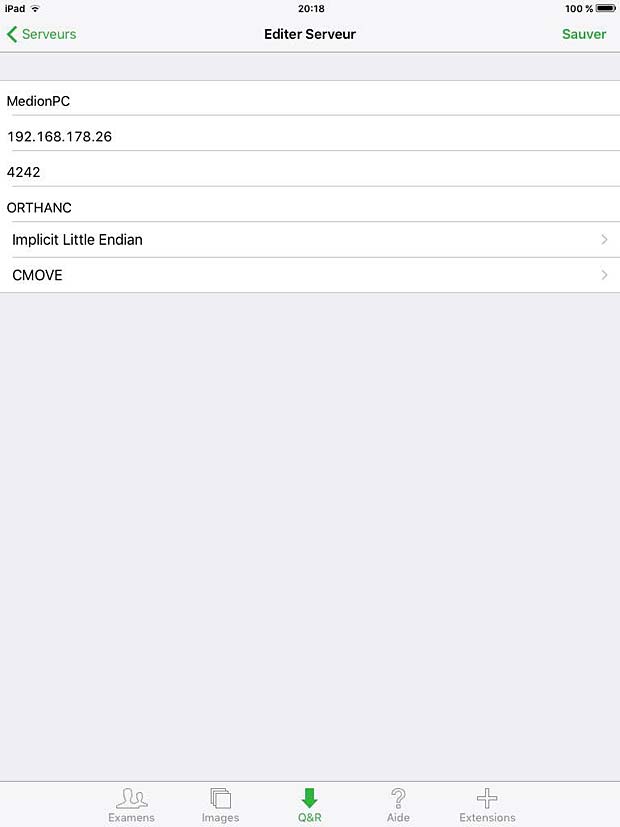
Osirix HD PACS Server configuration panel
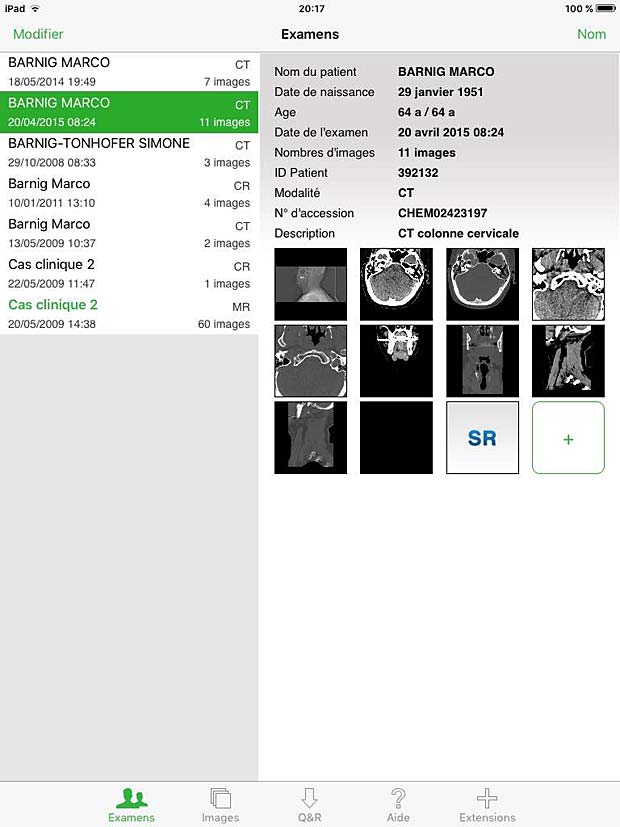
Osirix HD Exams Panel
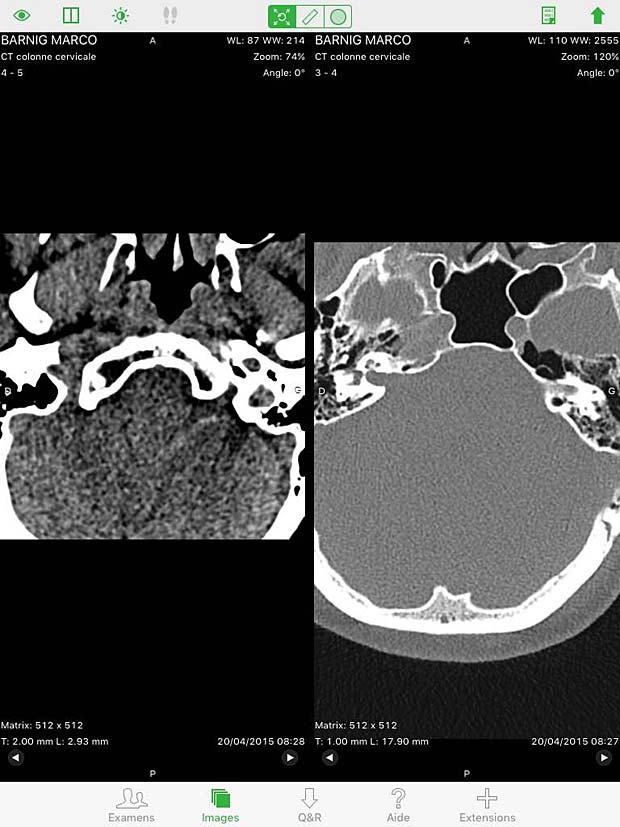
Osirix HD Split Screen in image viewer
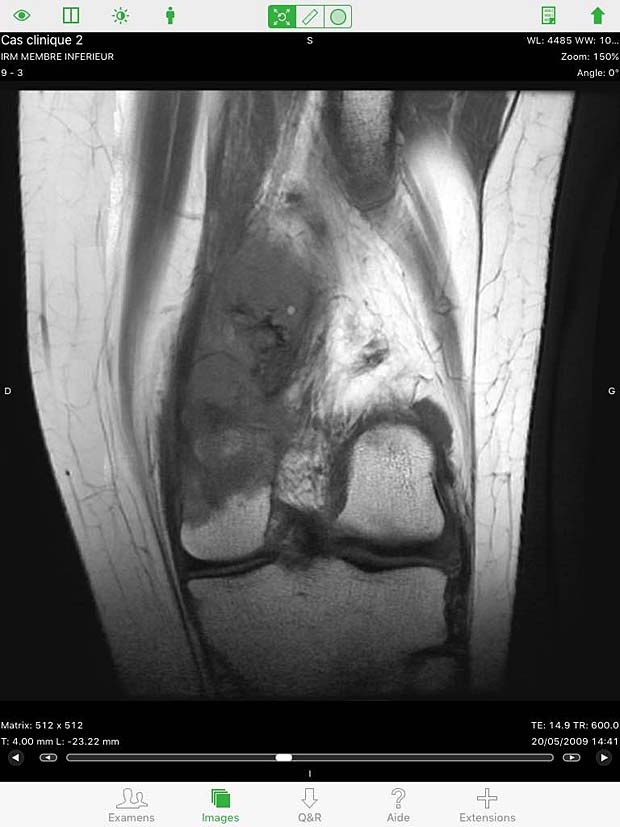
Osirix HD Image viewer with stack of 20 instances
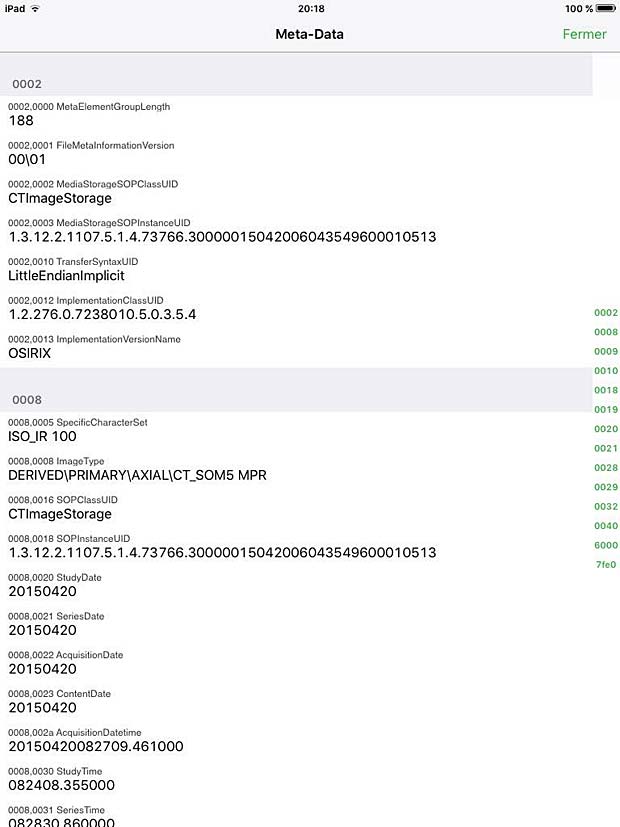
Osirix HD Metadata Panel
21. PACS DICOM Viewer
PACS DICOM Viewer app is a free, vendor-neutral PACS browser and DICOM viewer which works on iPad. It is developed by Logicpond Inc, a California based product design and development solutions company. The current version 2.1.1 was updated on November 11, 2015. The viewer can be configured to work with the Orthanc server, but the C-MOVE queries are aborted during the image transfer. The WADO protocol is also supported and works with Orthanc.
PACSDICOM Viewer startup picture
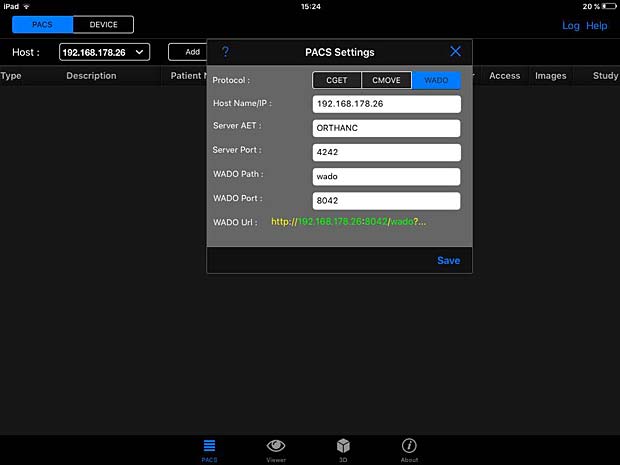
PACSDICOM Viewer configuration panel
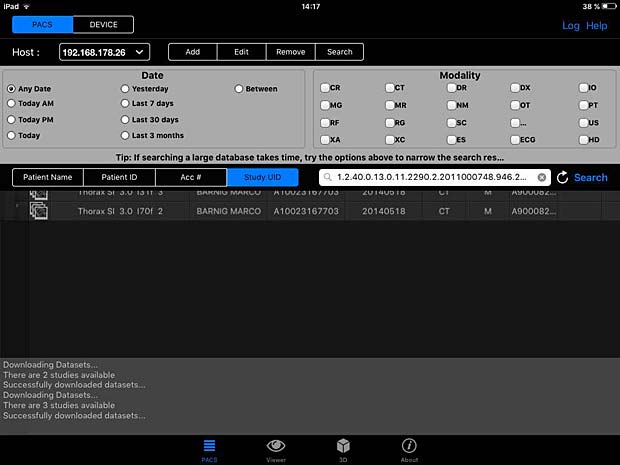
PACSDICOM Viewer search panel
PACSDICOM Viewer worklist panel
PACSDICOM Viewer download success
PACSDICOM Viewer images with rainbow LUT
PACSDICOM Viewer metadata panel
22. R-Site Mobile
R-Site Mobile for IOS supports 2D, 3D and MIP/MPR operations for DICOM files. To use this app, you have to own at least one set of R-Site PACS. R-Site is developed by DJ HealthUnion Systems Corporation in China. The current version 1.0.2.47 was updated on August 31, 2016.
23. ResolutionMD
ResolutionMD enables doctors to securely view patient images and reports from a wide variety of computers and mobile devices, collaborate with other practitioners and diagnose from any location. The product requires access to a configured ResolutionMD Server. It contains a demo mode to allow to experience the full functionality of ResolutionMD on a variety of anonymous patient images. ResolutionMD is devloped by Calgary Scientific Inc. The free version 5.3.3. was updated on July 19, 2016.
Android Versions
DICOM Android apps on Samsung Galaxy Tablet 2 – 7.0
The following iPad applications are also available as Android version on Google Play Store (in alphabetical order) :
3a. DICOM Mobile
The current Android version 1.3.2 of DICOM Mobile was updated on August 13, 2014. The price is the same as for the iOS version : 2.99$. The GUI and the features of the viewer are also identical to the iOS version. DICOM images can only be loaded from DropBox or from other web servers.
Android DICOM Mobile menu panel
9a. iPaxera
The Android free version 2.0.1 of iPaxera was updated on September 15, 2016. This app requires Android 4.4 or greater and I was not able to test it on my Samsung Galaxy tablet running Android 4.2.2. A few days later I changed my Blackberry phone with an LG Spirit phone running Android 5.0.1. After installing iPaxera on this phone, I noticed that the Android version is different from the iOS version and allows no PACS server configuration, but only dowloads of DICOM files from Dropbox or iTune sharing.
10a. iTA View
The Android free, but limited, version 1.10 of iTA View was updated on October 27, 2014. An in-app upgrade priced 26.99 EUR is available through the Google Play Store.
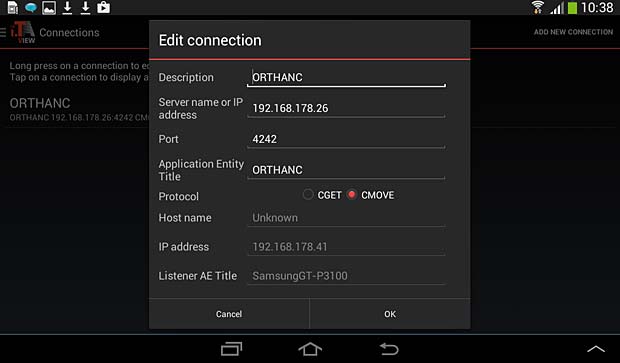
iTA View PACS server configuration panel
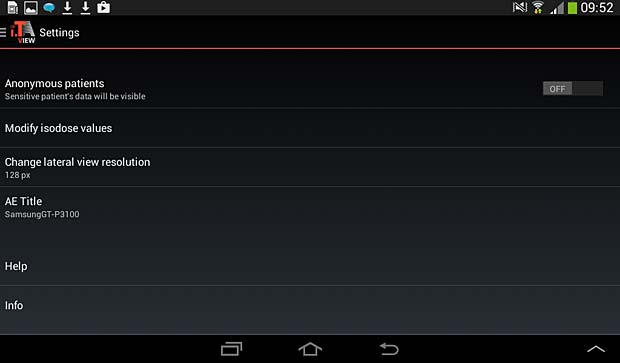
iTA View settings panel
iTA View search panel
12a. LEADTOOLS MedicalWebViewerDemo
The Android version of the LEADTOOLS MedicalWebViewer is the online live demo of the Medical Web Viewer Framework included with LEADTOOLS PACS Imaging and Medical Imaging Suite. The current version 1.4.1 was updated on February 2, 2014.
17a. mRay
The Android free version 4.1.6 of mRay was updated on September 23, 2016. The mRay app can not be used without the installation of the mRay Server Software.
Android Applications
The following DICOM viewers are only available as Android version (in alphabetical order) :
24. Dicomite
Dicomite is a free and powerful DICOM viewer with intuitive and simple user interface, optimized for use on multi-touch devices. Dicomite is developed by Samsung MNO Lab. The current version 1.7.1 was updated on October 11, 2016. To display images they must be downloaded into the local memory with an Internet browser.
DICOMITE image panel
25 Direct DICOM
The Direct DICOM Viewer is developed by Hui Zhang. The current version 1.2.4 was updated on March 21, 2015. To display images they must be downloaded into the local memory with an Internet browser. The app works however as DICOM server and local DICOM files can be stored to the Orthanc server.
Direct Dicom viewer image and metadata panels
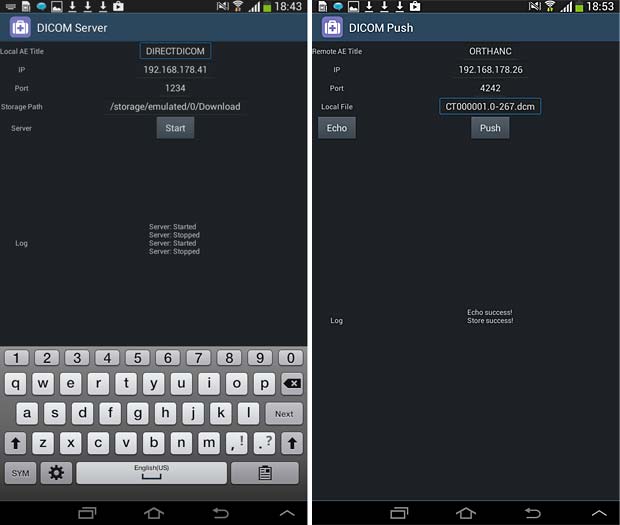
Direct Dicom viewer server and store (push) panels
26. Droid Dicom Viewer
The Droid Dicom Viewer was developed by Pierre Malarme. The free current version 3.2.3 was updated on May 17, 2011. Droid Dicom Viewer is a Free and Open Source Software (FOSS) licensed under the terms of the GNU General Public License. To display images they must be downloaded into the local memory (cache) with an Internet browser.
Droid DICOM viewer about panel

Droid DICOM viewer cache panel
Droid DICOM image panel
Droid DICOM viewer image panel with LUT/CLUT
27. DroidRender
DroidRender is a 3D DICOM viewer for Android, including DICOM file support by GDCM,
volume rendering and tissue segmentation and display. Developed by Startm, the current version 3.2.0 was updated on August 26, 2016. The app proposes in-app purchases at prices between €1.04 – €104.08 per item and displays ads at the bottom of the screen. To display images they must be downloaded into the local memory (cache) with an Internet browser.
Droid Render image panel
28. Mobile Dicom
Mobile Dicom is a free Android Mobile Dicom (Diconde) Viewer application which is based on opensource OFFIS DCMTK as its backend library. Inspired by opensource community and in the hope of extending usage of the library on Android devices, the Mobile Dicom / Diconde tool was created by Feng Gao. The current version 2.4 was updated on November 23, 2015. To display images they must be downloaded into the local memory (cache) with an Internet browser. The app works however as DICOM server and can store local DICOM files tthe Orthanc server.
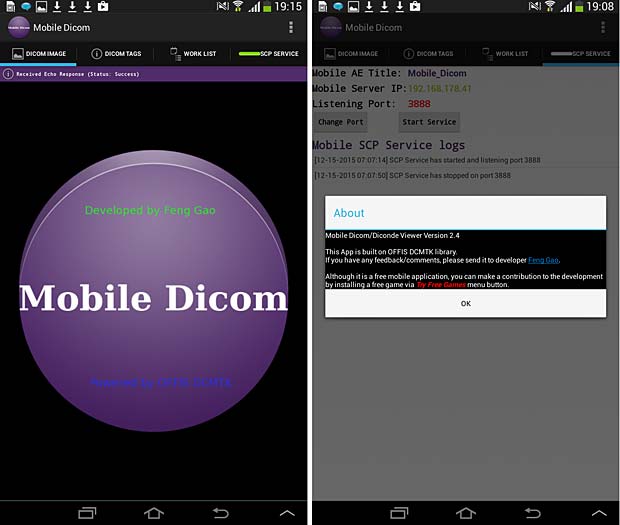
Mobile Dicom Viewer start and about panels
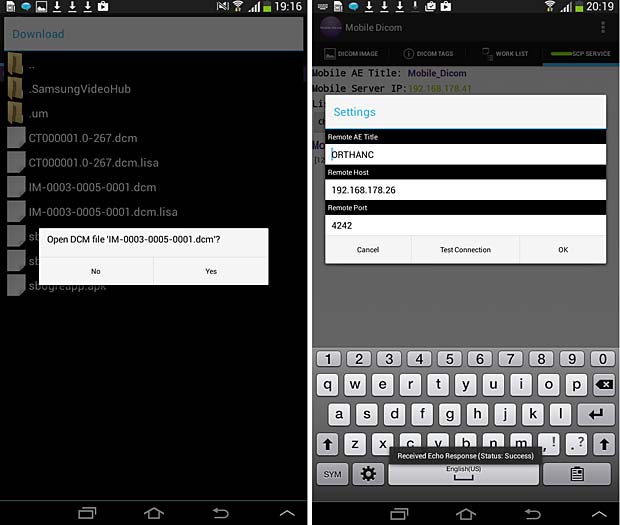
Mobile Dicom Viewer load and config panels; ECHO test was succesfull
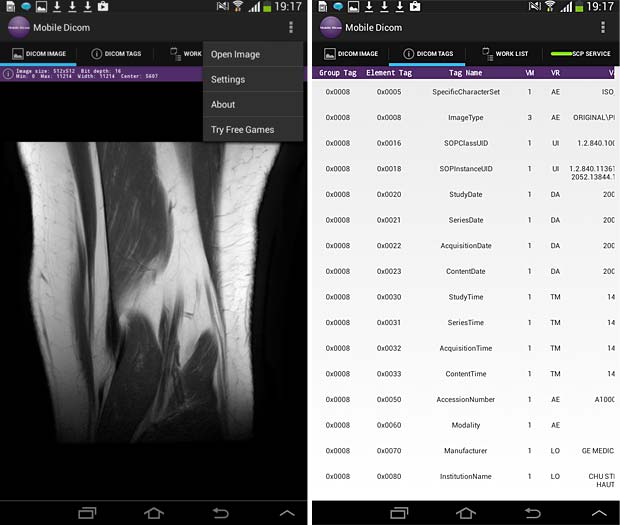
Mobile Dicom Viewer image and metadata panels
29. simplyDICOM
Simply DICOM is currently in public beta. It should work with any uncompressed DICOM images, downloaded into the local memory. The viewer has a minimal interface to keep the focus on the images themselves. The developer is Christopher Boyd. The current version 0.7.3 was updated on May 27, 2015. To display images they must be downloaded into the local memory (cache) with an Internet browser.
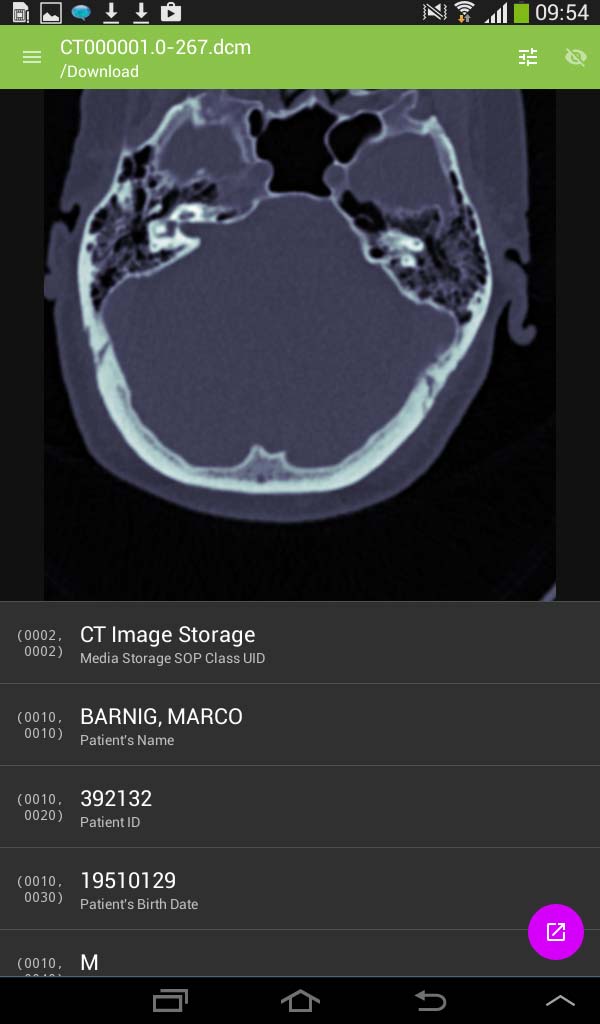
SimplyDICOM image viewer
DICOM file downloads
If DICOM files are served with the extension .dcm or/and with the content type application/dicom, they are recognized as DICOM images in iOS Safari and can be opened directly in the following DICOM viewers : MedFilm, MyXray, Osirix HD, DICOM IR, iMango or Dicom Mobile.

.dcm files recognized as DICOM images in iOS Safari browser by DICOM viewers
DICOM files embedded in .zip archives are recognized by the same DICOM viewers in the iOS Safari browser.

.zip files recognized in iOS Safari browser by DICOM viewers
On Android devices, DICOM files are only recognized by the DIRECT DICOM viewer in the Android browser, but they can be downloaded in the /storage/emulated/0/Download/ folder where they are accessible by most Android DICOM viewers.

Android DICOM file download panel
Links
The following list provides links to mobile applications which are not DICOM viewers, but related to medical imaging (in alphabetical order) :
- Aquarius Mobile (medical viewer, v 2.2, updated October 27, 2015, free)
- Centricity Radiology Mobile Access (medical viewer, v 4.0, updated December 1, 2013, free)
- DCMTK Command test (test tool, v 1.1.0, updated on September 11, 2015)
- iClarity (medical viewer, v 2.0.6, updated November 23, 2015, 4.99 $)
- JBJS Image Quiz (educational tool, v 1.1.3, updated January 15, 2014, 0.99 $)
- lifeIMAGE Mobile (educational tool, v 3.1.3, updated February 26, 2014, free)
- IMAIOS e-Cases (educational tool, v 1.3, updated February 2, 2014, free)
- IRM Prostate (educational tool, v 1.0, released August 21, 2012, 37.99 EUR)
- iSurf BrainView (educational tool, v 4.1.0, updated May 3, 2013, free)
- Medscape (educational tool, v 5.2.1, updated November 24, 2015, free)
- MOO Scans Bedtime Cases (educational tool, v 1.0.1, released December 28, 2012, free)
- NeuroPub Visualizer (medical viewer, v 1.2.2, updated June 13, 2013, free)
- PubMed On Tap (reference retrieval, v 2.8.1, updated July 30, 2015, 2.99 $)
- QEVLAR (education tool : quiz, v 1.1.2, updated October 16, 2014, free)
- RADIOLOGiQ apps (ten different apps for iPad and iPhone)
- Radiopaedia (educational tool, v 1.1, updated May 14, 2015, free)
- ResolutionMD (medical viewer, v 5.1.3, updated November 19, 2015, free)
- Surgical Radiology (educational tool, v 1.0, released May 13, 2011, free)
Conclusions
Among the 29 tested native mobile DICOM viewers, 18 are iOS apps, 6 are Android apps and 5 are available on the two platforms. 9 viewers can be configured to access a vendor-neutral PACS server, but only 5 of them work as expected and comply with the Orthanc PACS server. Among them are the three renowned commercial viewers iMango, iTA View and Osirix (priced between 15 and 50 USD), one free app (iPaxera) developed by a world leading medical imaging solution provider (Paxeramed Corp) and one app (DICOM IR) created by an independent free-lance developer ( Pietro Zuco). The other 4 configurable viewers are limited, partially disabled or crashing.
The remaining viewers are demo apps working with proprietary PACS systems, displaying only DICOM files loaded from DropBox or from another web server or crashing and working not at all. There are additional DICOM viewer apps in the AppStore and the Google PlayStore which are not listed because they were not updated in the last two years.
My two cents
I was shocked to see that most native mobile apps in the medical domain are not state of art. Apps in the stores which have not been updated in the last few years, text errors in the menus, program errors in the logic ( 2 viewers change the AETitle during communications), crippled GUI’s when the orientation of the device changes, wrong version numbers in the about panel etc, are testimonies of the poor quality.